Decoding the molecular basis of mental illness
It is the mission of my group to develop novel paradigms to understand the genetic basis of complex psychiatric diseases such as schizophrenia and bipolar disorder. To implement this goal, the lab for Functional Genomics in Psychiatry pursues a combined experimental and computational strategy to deconstruct the polygenic risk architecture underlying complex diseases with the goal to decode the molecular mechanisms that contribute to disease onset and progression and treatment resistance.
Moreover, it is the central goal of my lab to rapidly operationalize these insights to optimize patient treatment, obtain insights into the molecular cause of treatment resistance and identify new drug targets.
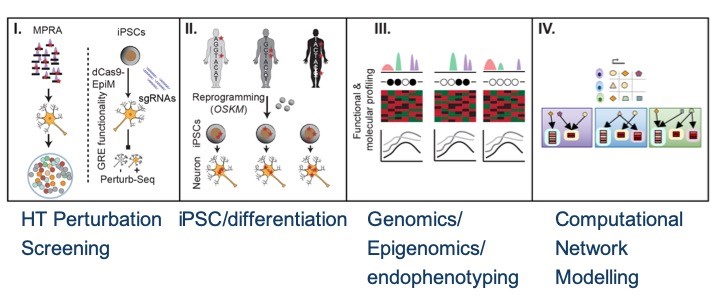
The lab for Functional Genomics in Psychiatry leverages cutting edge statistical and deep learning approaches, pluripotent stem cell based models, functional genomics and patient derived medical record data to empower personalized medicine in practice. In particular, we i.) develop and apply novel computational strategies to decode the personalized functional consequences of common disease associated genetic variants in individual patients, utilizing multidimensional –omics data types of large cohorts (e.g. UKBiobank) and machine learning, ii.) utilize standardized human induced pluripotent stem cell derived neural cell populations as a versatile platform to model the molecular and cellular polygenic basis of these diseases and test the computational predictions, iii.) employ high throughput genomic profiling using a variety of assays including ChIP-Seq, ATAC-Seq, HiC/capture HiC, single cell RNA-Seq/ATAC-Seq & SNP arrays of in vitro derived and primary, post mortem cell populations iv.) develop and apply different types of high-throughput functional genomic screening technologies such as massively parallel reporter assays (MPRA), shRNA/sgRNA as well as Perturb-Seq to functionally characterize (combinations) of disease associated non-coding gene regulatory elements in different in vitro derived human cell types and v.) apply machine learning techniques to mine electronic health records in order to connect molecular insights to patient (endo-)phenotypes.In this context, we closely collaborate with multiple clinical groups and large established research cohorts for patient recruitment and clinical characterization to apply our aforementioned analysis pipeline to distinct patient strata defined based on clinical and genetic parameters such as polygenic risk, disease course or treatment response.
It is our main goal to rapidly operationalize the molecular and neurobiological insights emerging from our experiments in order to effectively implement a workflow starting from the bedside to the bench and back.